Research Areas
Our computational biology and disease genomics lab is focused on developing computational methods, in close collaboration with experimental scientists, to uncover how genetic variations contribute to human disease and accelerate the discovery of novel therapeutic targets.
Genetic Basis of Human Diseases
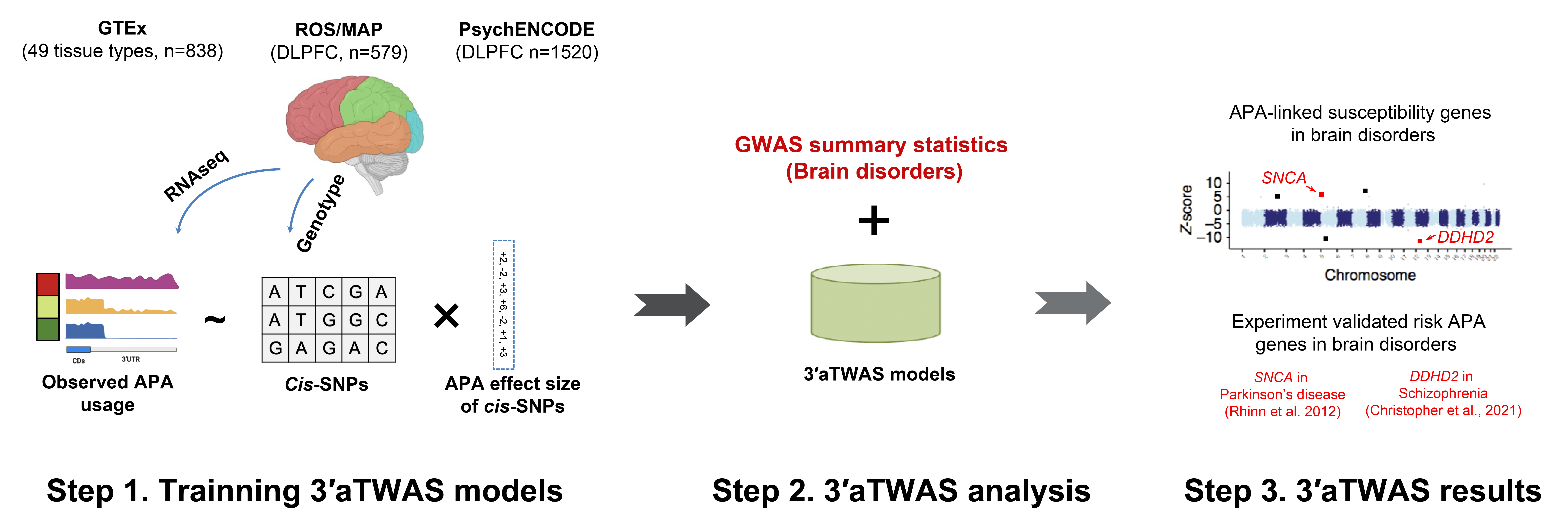
Our research aims to dissect the genetic and molecular mechanisms underlying human disease risk by integrating large-scale genomic and multi-omic data. We firstly focus on alternative polyadenylation (APA). APA occurs in approximately 70% of human genes and substantively impacts cellular proliferation, differentiation, and tumorigenesis. Mechanistically, 3’aQTLs could alter polyA motifs and RNA-binding protein binding sites, leading to thousands of APA changes. We described the first atlas of human 3’-UTR alternative polyadenylation (APA) Quantitative Trait Loci (3’aQTLs) and developed a new method 3′ UTR APA TWAS (3′aTWAS) to indentify diseases risk APAs (JCI 2025, Nature Communications 2023, Nucleic Acids Research 2022, Nature Genetics 2021).
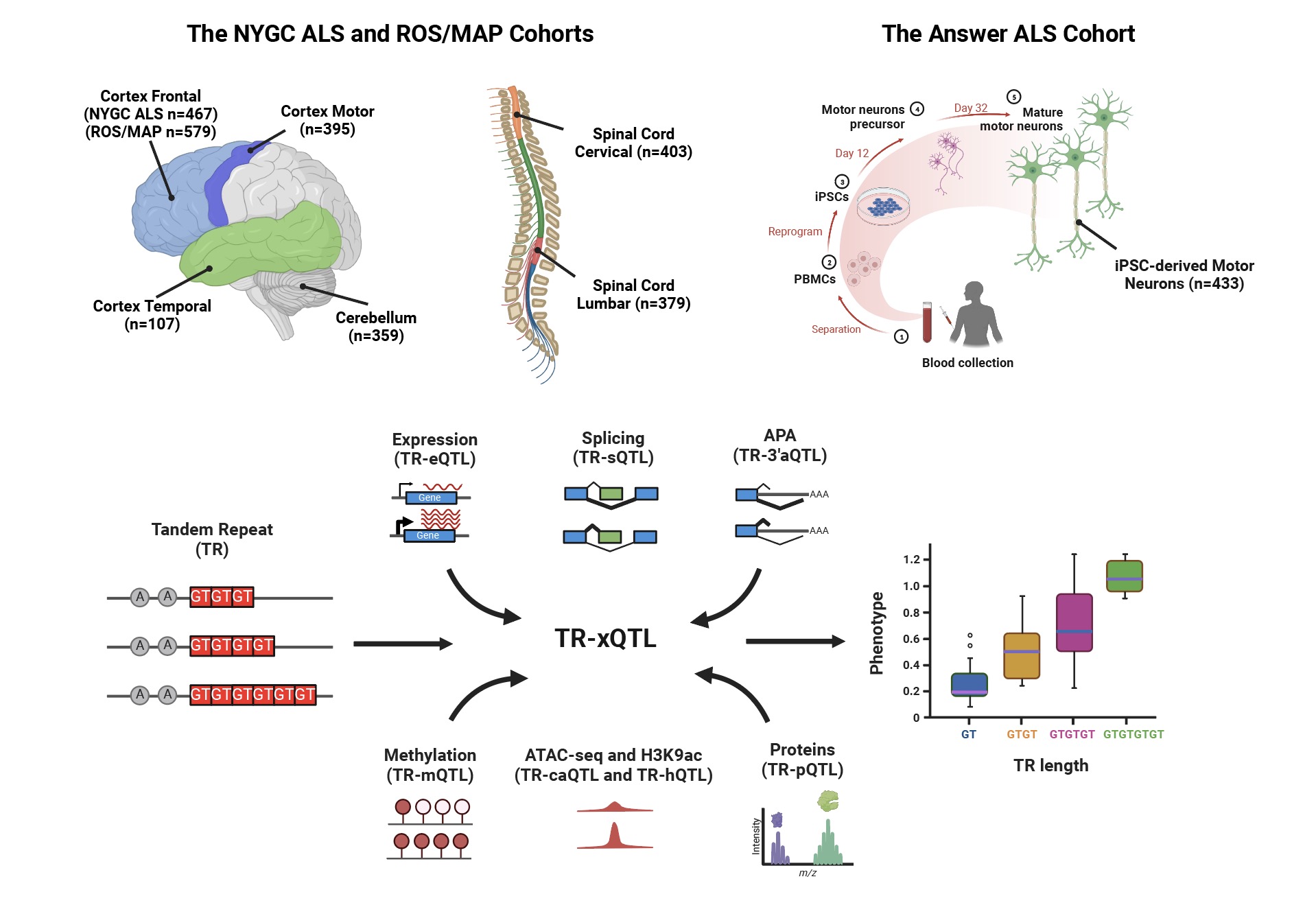
Human Repeatome and Diseases
Tandem Repeats (TRs), i.e., sequences where a TR unit or motif (e.g., CAG) is consecutively repeated like CAG-CAG-CAG, comprise about 8% of the human genome, spanning millions of loci in both coding and noncoding regions. Recent case studies have demonstrated that TR variations, through either expansions or contractions, lead to changes in the number of TR units and corresponding TR lengths, significantly impacting coding sequences, gene expression or splicing. These changes have been linked to increased risks of ~60 human diseases, primarily identified through family-based linkage analyses. Notable examples include amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), Huntington’s disease, and multiple cancers. Recently, we developed the Tandem Repeat Aggregation Atlas (TR-Atlas), a biobank-scale reference of 0.86 million TRs derived from 338,963 whole genome sequencing (WGS) samples of diverse ancestries (39.5% non-European samples) (Cell, 2024). We also investigated how TR variations affecting genes regulation in brain tissues and increasing brain disease risks (Nature Genetics, 2025).